Segminator II now contains a feature for the
probabilistic
read
error
detection
across
temporally
obtained
reads
(PREDATOR).
In brief, the approach utilizes joint nucleotide frequencies across temporally sampled data sets from a single
host, in conjunction with known platform error rates, to derive a set of posterior probabilities for each of the
possible nucleotides at each site. Posterior values indicate the probability of a nucleotide observed within any of
the individual data sets, regardless of frequency, being biologically relevant to the viral population and, thus,
not being due to platform error.
Error correction then involved the replacement of individual nucleotides associated with low posterior probabilities
with the population consensus; thus avoiding the need to discard poor quality reads.
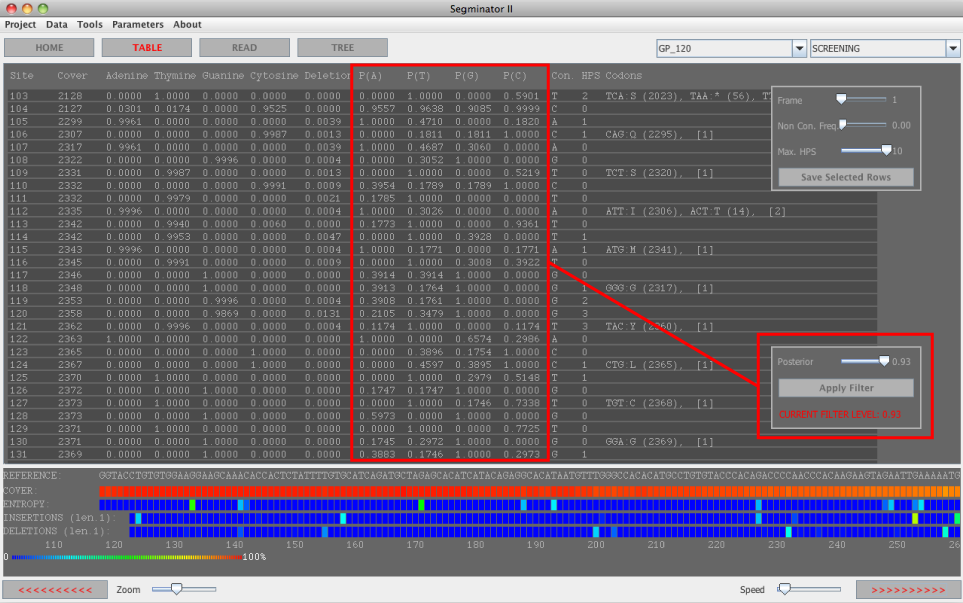
Figure 1:Predator within Segminator II
The sole requirement for the application of PREDATOR to the data is that more than one dataset is associated with the project. When
this is the case the boxed area in figure 1 becomes active and the user can select a correction posterior.
Once a posterior threshold has been
selected all reads, trees and base frequencies within the software will reflect correction based on the value chosen. To undo the correctional procedure the slider can simply be set to 0 - indicating that all variants with a posterior of 0 or more will be used.
Updates
5th July, 2012
free for academics | GNU Lesser GPL licence | software not guaranteed


