Integrated network analysis of expression data and disease phenotypes
Transcriptomics technologies such as RNA-sequencing and microarray are invaluable in the study of human disease by identifying differentially expressed genes between experimental groups. A common approach to analyse these data is to overlay the expression information onto a protein-protein interaction (PPI) network. However, the resulting generic network lacks biological context and contains interactions not relevant to the biological condition under study. Further integration of disease related information could guide identification of the most biologically important regions of the interactome with regards to a disease transcriptomics dataset.
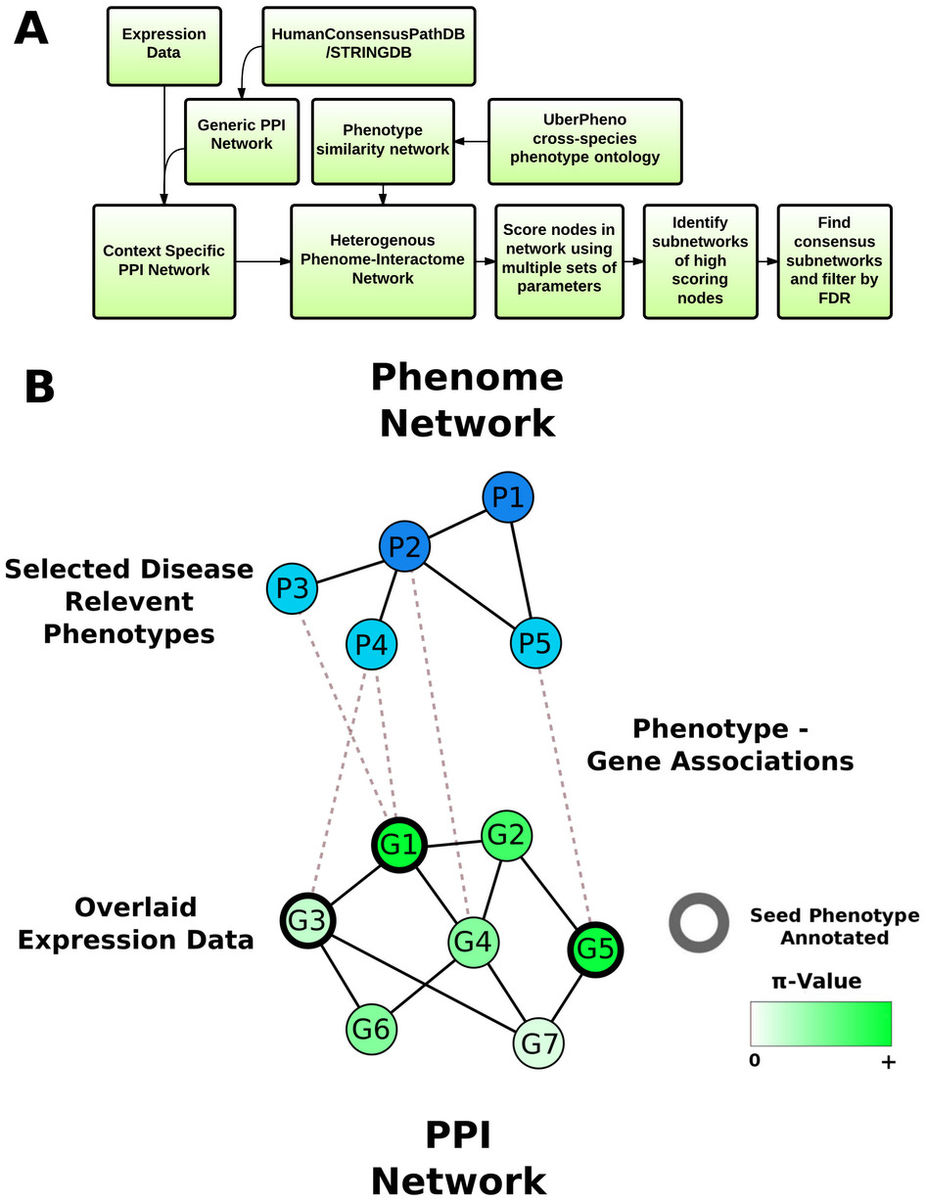
We created a tool, PhenomeExpress, that utilises both the transcriptomics data being analysed and the prior knowledge of all cross-species phenotype to gene associations, including those directly related to the disease under study. We adapted and combined existing algorithms for node ranking and sub-network detection with a cross-species phenome-interactome network to automatically detect sub-networks of proteins from transcriptomics data that are relevant to the disease of interest.
Downloads
- Source code of PhenomeExpress: https://github.com/soulj/PhenomeExpress
- Cytoscape app (PhenomeScape): http://apps.cytoscape.org/apps/phenomescape
References
Soul J, Dunn SL, Hardingham TE, Boot-Handford RP, Schwartz JM (2016). PhenomeScape: a Cytoscape app to identify differentially regulated sub-networks using known disease associations. Bioinformatics 32: 3847-9.
PubMed | Full text
Soul J, Hardingham TE, Boot-Handford RP, Schwartz JM (2015). PhenomeExpress: A refined network analysis of expression datasets by inclusion of known disease phenotypes. Scientific Reports 5: 8117.
PubMed | Full text