Using pathway networks to model context dependent cellular function
Molecular networks are commonly used to explore cellular organisation and disease mechanisms. Function is studied using molecular interaction networks, such as protein-protein networks. Functional modules can be identified based on molecular network topology, however these networks struggle to depict the temporal nature of interactions, giving the impression that all interactions are constant. The imperfect nature of experimental protein interaction data further impedes functional module extraction. Representation of genes by single nodes artificially merges the functions of pleiotropic genes, distorting the arrangement of function within molecular networks.
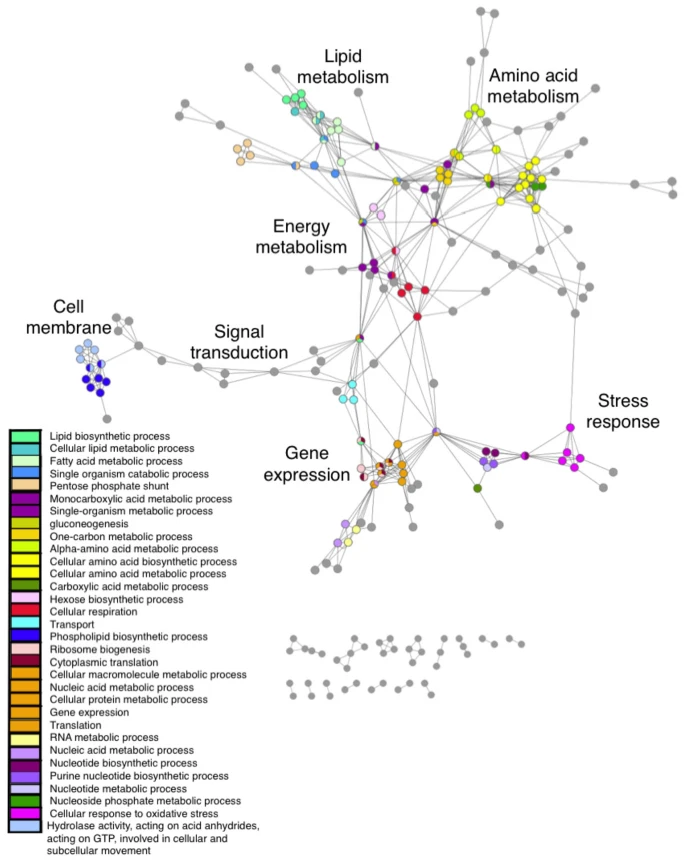
Pathways are composed of sets of proteins that are known to interact within a particular cellular context, corresponding to a discernible biological function. Their representation of context dependent cellular activity makes them ideal for use as nodes within a new pathway level model. Using combinatorial algorithms, we produced a reduced redundancy pathway set to represent global cellular systems. Enrichment analysis provides reliable functional annotations for each pathway node, attributing independent functions to pleiotropic genes. Edges are based on functional semantic similarity, generating a network representation of functional organisation.
The pathway model provides a complementary, high-level functional model that begins to bridge the gap between molecular data and phenotypes. The utilisation of pathway data provides a large, well-validated data source, avoiding the inaccuracies inherent with molecular data. Pathway models better represent components of biological complexity such as pleiotropy and linear implementation of functions.
References
Stoney RA, Schwartz JM, Robertson DL, Nenadic G (2018). Using set theory to reduce redundancy in pathway sets. BMC Bioinformatics 19: 386.
PubMed | Full text
Stoney R, Robertson DL, Nenadic G, Schwartz JM (2018). Mapping biological process relationships and disease perturbations within a pathway network. npj Systems Biology and Applications 4: 22.
PubMed | Full text
Stoney R, Ames R, Nenadic G, Robertson DL, Schwartz JM (2015). Disentangling the multigenic and pleiotropic nature of molecular function. BMC Systems Biology 9(Suppl 6): S3.
PubMed | Full text