Modelling the Neurospora circadian clock and its response to light and temperature
Circadian clocks are internal timekeepers that aid survival by allowing organisms, from photosynthetic cyanobacteria to humans, to anticipate predictable daily changes in the environment and make appropriate adjustments to their cellular biochemistry and behaviour. Computational modelling has aided our understanding of the molecular mechanisms of circadian clocks, nevertheless it remains a major challenge to integrate the large number of clock components and their interactions into a single, comprehensive model that is able to account for the full breadth of clock properties.
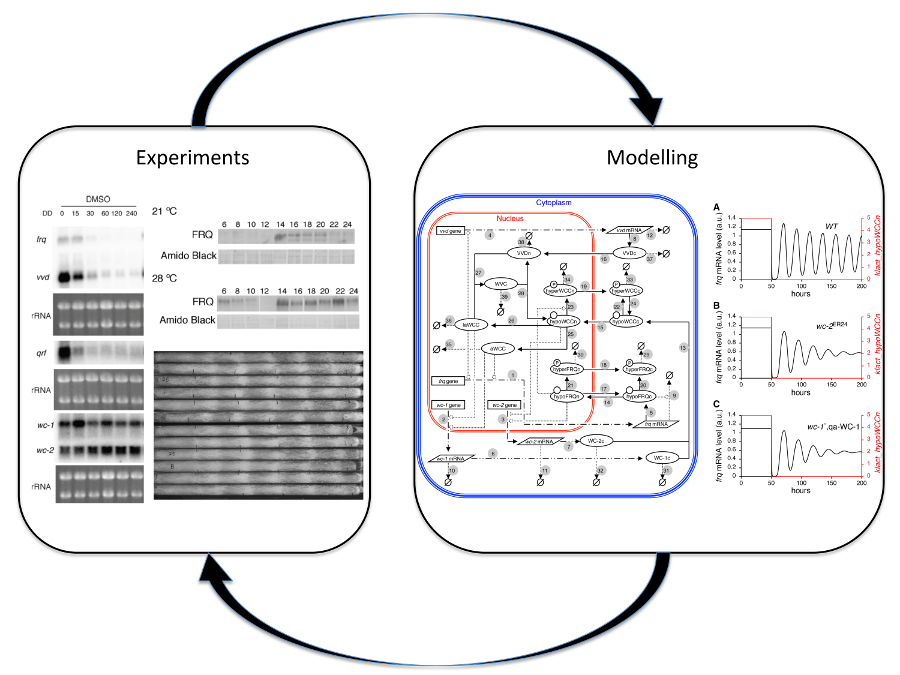
We have constructed a comprehensive dynamic model of the Neurospora crassa circadian clock that incorporates its key components and their transcriptional and post-transcriptional regulation. Simulations from the model identified components that strongly influence the period and amplitude of the molecular oscillations and predicted the possible key mechanisms allowing the clock to respond to environmental cues, such as light and temperature. Experiments have been carried out to validate the predictions from the model and simulations were made to explore possible mechanisms. The dry (modelling) and wet (experimental) results inform each other and bring about efficient and cost-reducing research in this typical systems biology approach.
Download
Circadian clock model of Neurospora crassa available from the BioModels database: BIOMD0000000437
Reference
Tseng YY, Hunt SM, Heintzen C, Crosthwaite SK, Schwartz JM (2012). Comprehensive modelling of the Neurospora circadian clock and its temperature compensation. PLoS Computational Biology 8: e1002437.
PubMed | Full text