GRaPe – Gene Reaction Protein Builder
Despite considerable advances in the topological analysis of metabolic networks, inadequate knowledge of enzyme kinetic rate laws and their associated parameter values still hampers large-scale kinetic modelling. Furthermore, the integration of gene expression and protein levels into kinetic models is not straightforward.
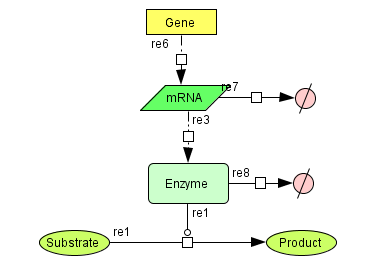
The GRaPe software was developed to enable the construction of kinetic models for large systems of metabolic reactions and associated gene expression processes. It generates generic rate equations for all reactions in a model and encompasses two robust methods for parameter estimation from time series of 'omics' data. It also allows for the seamless integration of gene expression and protein levels into a reaction and generates equations for both transcription and translation.
Downloads
- Download GRaPe – Requires Java JRE 6 or higher
- How to install and User guide
- Example files: Glycolysis model with varying glucose level (SBML L2V1) and Input data for parameter estimation.
References
Adiamah DA, Schwartz JM (2012). Construction of a genome-scale kinetic model of Mycobacterium tuberculosis using generic rate equations. Metabolites 2: 382-97.
Full text
Adiamah DA, Handl J, Schwartz JM (2010). Streamlining the construction of large-scale dynamic models using generic kinetic equations. Bioinformatics 26: 1324-31.
PubMed | Full text