


Next: Refinements to substitution models
Up: Base-paired substitution models implemented
Previous: RNA7D model (Tillier, 98)
Contents
PHASE
contains a general 16-state model (RNA16), however this model has 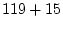 free parameters
and is not well suited for phylogenetic inference, especially maximum-likelihood inference.
RNA16A is a simplified 16-state model, it reduces some of the complexity of the RNA16
model by cutting down on the number of rate parameters from 120 to 5. It uses a rate
of single transitions
free parameters
and is not well suited for phylogenetic inference, especially maximum-likelihood inference.
RNA16A is a simplified 16-state model, it reduces some of the complexity of the RNA16
model by cutting down on the number of rate parameters from 120 to 5. It uses a rate
of single transitions 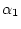 , a rate of double transversions
, a rate of double transversions 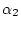 , a
mismatch
, a
mismatch
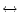 non-mismatch transition rate
non-mismatch transition rate 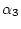 for transitions
requiring only one substitution and a mismatch
for transitions
requiring only one substitution and a mismatch
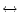 mismatch transition
rate
mismatch transition
rate 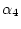 for transitions requiring one substitution too. The reference rate is the rate of double transitions.
Some base-pair substitutions are not allowed (null substitution rate).
The transition matrix for the RNA16A model is given in
table 2.10.
for transitions requiring one substitution too. The reference rate is the rate of double transitions.
Some base-pair substitutions are not allowed (null substitution rate).
The transition matrix for the RNA16A model is given in
table 2.10.
Table 2.10:
RNA16A transition matrix
 |
Gowri-Shankar Vivek
2003-04-24
