

Next: Base-paired substitution models implemented
Up: Paired-site substitution models
Previous: RNA secondary structure
Contents
Theory of compensatory substitutions
From the individual sequence viewpoint complementary mutations
are a two-step process typically involving a U-G or a
G-U pair as a transition state. These pairs are thermodynamically
less stable than Waston-Crick pairs but they are still more likely to arise
than any other mismatches. Nonetheless, in phylogenetic studies we
are not considering individual copies of a gene but we are rather
modelling consensus sequences for a large number of individuals. From
the population genetics viewpoint, evolution in stems can either
occur by two single substitutions or by simultaneous
compensatory substitutions, (see, e.g.,
Savill et al., 2001; Higgs, 1998).
The first mechanism is by fixation of the slightly deleterious UG or GU
pair in the population before the second mutation occurs. The second
mechanism happens when natural selection against intermediate mutants
is too strong. In such a case, deleterious pairs are kept low in
frequency until a second mutation takes place in one of the
sporadic mutant sequences by chance. Afterwards, the new neutral
variant may replace the original one due to drift in gene
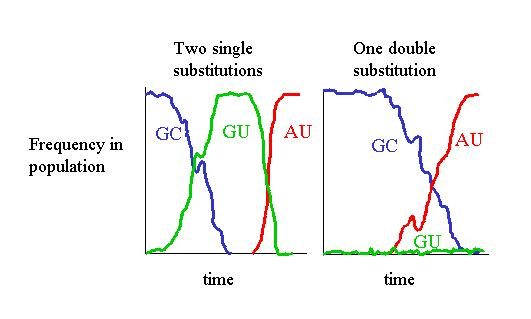
frequencies (see figure 2.4).
Figure 2.4:
Substitution mechanisms for paired-sites
|
|
Therefore, even if simultaneous mutations are very unlikely to
occur in a single organism, it is reasonable, although not
compulsory, to allow double substitutions in models from the
population point of view. The experimental results you can have with PHASE
confirm that. Since natural selection against intermediate mutants with any
other mismatch pairs than U-G or G-U is usually much stronger,
one can notice two groups of states in which rapid interchange occurs, while
interchange between the two groups,
although possible, is really slow (see figure 2.5).
Figure 2.5:
Mutation rate between paired-sites
|
|



Next: Base-paired substitution models implemented
Up: Paired-site substitution models
Previous: RNA secondary structure
Contents
Gowri-Shankar Vivek
2003-04-24