


Next: Using consensus
Up: mcmcphase
Previous: mcmcphase
Contents
The mcmcphase program perfoms
Bayesian estimation of phylogenies and uses Markov chain Monte Carlo to produce
large samples from the posterior probability density.
To use mcmcphase, simply type at the command-line:
mcmcphase mcmcphase-control-file
where mcmcphase-control-file is a valid control file for
the mcmcphase program.
The mcmcphase program saves the results of an inference in many files.
Be warned that it might require a large amount of disk space for large studies
(around 90 Mb for 70 species and 50000 samples).
 .besttree and .bestmodel files:
.besttree and .bestmodel files:
- the phylogeny and the parameters of the
substitution model when the best state (i.e.,
the state with the highest
likelihood) was visited, it is not necessary one of the sampled configurations
and this state might have been visited during the burnin period.
The best configuration is not very important in a MCMC analysis but a ``strange'' best state
indicates quickly that something went wrong. The tree and the model can also
be used as starting points in maximum likelihood inference.
 .mp file:
.mp file:
- the file with the sampled parameters of the substitution model(s). Each sample
occupies one line. The parameters are, in order,
 the proportion of invariant sites if an invariant category is used (+I models)
the proportion of invariant sites if an invariant category is used (+I models)
 the gamma shape parameter (
the gamma shape parameter (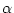 ) if the discrete gamma model is used (+dGX models)
) if the discrete gamma model is used (+dGX models)
 the frequencies of the states as they appear in the substitution matrix
the frequencies of the states as they appear in the substitution matrix
 the rate ratios
the rate ratios
When a MIXED model is used, substitution model parameters are printed
sequentially. Except for the first model, each set of parameters is preceded
by the average substitution rate of the model. The average substitution rate
for the first model is always 1.0 and therefore this value is not reported.
 .samples file:
the sampled topologies, this file can be used with another phylogenetic package
to produce a consensus tree. To avoid wasting disk space, mcmcphase will
output the sampled topologies using an index for each species according to their
appearance order in the datafile.
.samples file:
the sampled topologies, this file can be used with another phylogenetic package
to produce a consensus tree. To avoid wasting disk space, mcmcphase will
output the sampled topologies using an index for each species according to their
appearance order in the datafile.
 .bl file:
the branch lengths for the previous topologies (for use with
other PHASE
programs).
.bl file:
the branch lengths for the previous topologies (for use with
other PHASE
programs).
 .output file:
a file with similar content to the screen output.
.output file:
a file with similar content to the screen output.
 .plot file:
the evolution of the likelihood during the run. Sampling of these
values starts at the beginning of the run, i.e.,
likelihood values are stored
durning the burnin too.
.plot file:
the evolution of the likelihood during the run. Sampling of these
values starts at the beginning of the run, i.e.,
likelihood values are stored
durning the burnin too.



Next: Using consensus
Up: mcmcphase
Previous: mcmcphase
Contents
Gowri-Shankar Vivek
2003-04-24
 the proportion of invariant sites if an invariant category is used (+I models)
the proportion of invariant sites if an invariant category is used (+I models)
 the gamma shape parameter (
the gamma shape parameter (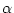 ) if the discrete gamma model is used (+dGX models)
) if the discrete gamma model is used (+dGX models)
 the frequencies of the states as they appear in the substitution matrix
the frequencies of the states as they appear in the substitution matrix
 the rate ratios
the rate ratios